sharing, publishing, archiving
Karen Cranston, Hilmar Lapp
URL to lesson:
https://github.com/Reproducible-Science-Curriculum/rr-publication

To the extent possible under law,
the person who associated CC0
with this work has waived all copyright and related or neighboring
rights to this work.
This work is published from:
United States.
Non synomymous
sharing, publishing, archiving
- shared could mean I emailed it to you ;)
- publish : citable artifact, discoverable
- archive : long-term preservation
why, what, when, where, how, with whom to share & publish?
- why publish, share, archive?
- what materials do we need to publish?
- when do we make them available?
- who are we sharing with?
- where do we publish various outputs?
- how do we prepare materials for publication?
For the remaining slides we are going to assume that we are at the point of publication.
why publish, share, archive?
Write down your top 3 reasons. When prompted, compare with your neighbor(s).
Consider:
- Mandates
- Norms
- Value for you
- Value for science
why?
- funding agency / journal requirement
- community expects it
- increased visibility / citation
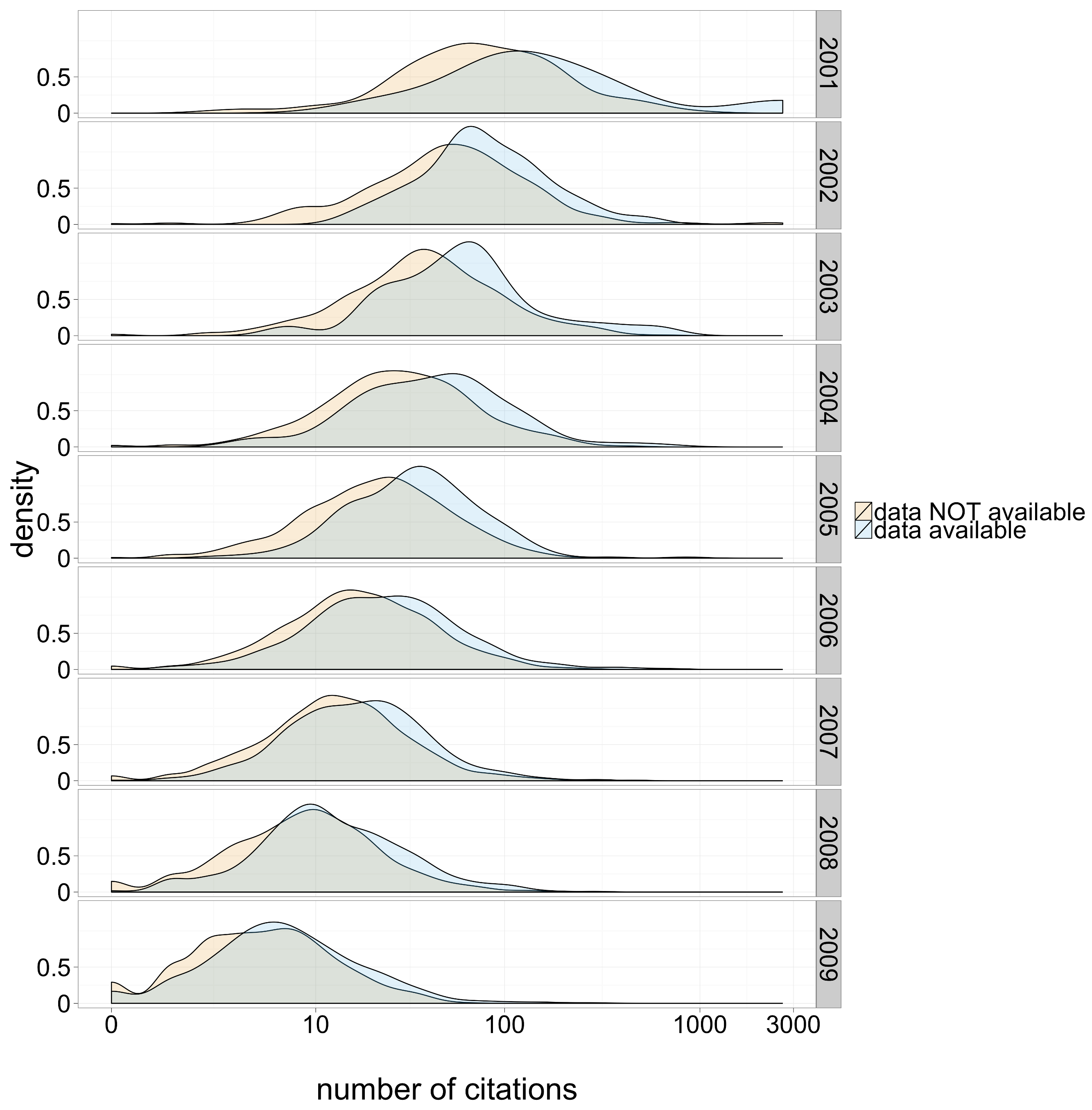
increased visibility / citation
Piwowar & Vision (2013) “Data reuse and the open data citation advantage.” PeerJ, e175
Figure 1: Citation density for papers with and without publicly available microarray data, by year of study publication.
why?
- funding agency / journal requirement
- community expects it
- increased visibility / citation
- better research
better research
Wicherts et al (2011) “Willingness to Share Research Data Is Related to the Strength of the Evidence and the Quality of Reporting of Statistical Results.” PLoS ONE 6(11): e26828
Figure 1. Distribution of reporting errors per paper for papers from which data were shared and from which no data were shared.
why?
- funding agency / journal requirement
- community expects it
- increased visibility / citation
- better research
- higher quality
- more efficient, less redundant science
Activity (in pairs)
Catalog the artifacts you produced this morning.
- What needs to be published, and why?
- What does not need to be published, and why?
- Anything that cannot be published?
it depends
share? yes!
- starting data set
- metadata
- data cleaning steps
- analysis scripts
- source code
- readme
share? maybe?
- raw data
- processed / cleaned data
- intermediate results
share? no!
- confidential (e.g., patient) data
- material already published
- pre-existing restrictive license
- passwords, private keys
where?
how to choose?
- is there a domain specific repository?
- what are the backup & replication policies?
- is there a plan for long-term preservation?
- can people find your materials?
- is it citable? (does it provide DOIs)
- is your purpose archival, sharing or publication?
what goes where when?
You will likely have different artifacts:
- Rmarkdown
- source code
- other documentation
- raw data
- derived data
Possible workflow:
- develop data & code on GitHub
- upon publication
- share markdown on RPubs
- archive a snapshot of data in Dryad
- code snapshot to Zenodo
how to share, publish: file formats
Do's
- non-proprietary file formats
- text file formats (.csv, .tsv, .txt)
Don't's
- proprietary file formats (.xls)
- data as PDFs or images
- data in Word documents
how to share, publish: checklist
- top-level
READMEthat describes the data or software package - list files and naming conventions
- describe abbreviations, column names, etc
- installation and usage instructions for software
- create separate
INSTALLif long
- create separate
- citation instructions
- consider creating a
CITATIONfile
- consider creating a
- contribution instructions
- Github will automatically link to
CONTRIBUTINGfile for new issues and pull requests
- Github will automatically link to
Activity (in pairs)
Documenting your research:
- collect all of the to-be-archived artifacts from the preceding lesson into a directory
- write a README file that describes the contents of the directory
Put a license or waiver on it
does copyright apply?
Copyright applies to creative works
- source code
- text (manuscripts etc)
- images
Typically not copyrightable:
- data, results
- individual records in a database of facts
Depends on jurisdiction and case:
- curated collections of data?
- databases
- medical images?
Choose A License
software licensing guide
Morin, Andrew, Jennifer Urban, and Piotr Sliz. 2012. “A Quick Guide to Software Licensing for the Scientist-Programmer.” PLoS Computational Biology 8 (7): e1002598.
open is not open to interpretation
The Open Definition sets out principles that define “openness” in relation to data and content. It makes precise the meaning of “open” in the terms open data, open content, and open source:
“Open means anyone can freely access, use, modify, and share for any purpose (subject, at most, to requirements that preserve provenance and openness).”
or more succinctly:
“Open data and content can be freely used, modified, and shared by anyone for any purpose”
Waiving copyright

CC0 enables scientists, educators, artists and other creators and owners of copyright- or database-protected content to waive those interests in their works and thereby place them as completely as possible in the public domain, so that others may freely build upon, enhance and reuse the works for any purposes without restriction under copyright or database law.
Dryad requires CC0
Dryad’s use of CC0 to make the terms of reuse explicit has some important advantages:
- Interoperability: Since CC0 is both human and machine-readable, other people and indexing services will automatically be able to determine the terms of use.
- Universality: CC0 is a single mechanism that is both global and universal, covering all data and all countries. It is also widely recognized.
- Simplicity: There is no need for humans to make, or respond to, individual data requests, and no need for click-through agreements. This allows more scientists to spend their time doing science.
licenses versus community norms
From the Panton Principles:
[…] in the scholarly research community the act of citation is a commonly held community norm when reusing another community member’s work.
Community norms can be a much more effective way of encouraging positive behaviour, such as citation, than applying licenses. A well functioning community supports its members in their application of norms, whereas licences can only be enforced through court action and thus invite people to ignore them when they are confident that this is unlikely.
licenses are legal instruments
- Licenses, copyright, terms of use are complicated issues.
- There are legal implications to your choices.
- Citation is a professional norm in science.
- We have good systems for ensuring proper citation.
- Would you try to sue someone in court who fails to cite you properly?
- Keep it simple by putting the least-restrictive license possible
Let scientists do science without having to talk to lawyers.